Vessel segmentation in brain 3D DSA(Dyna CT) and MRA image is the first step of the project.
It is quite fortunate that the image quality is ideal, so the auto grayscale in Matlab imadjuct function is good enough to segment vessels.
The effect of segmentation and code for segmenting listed below.
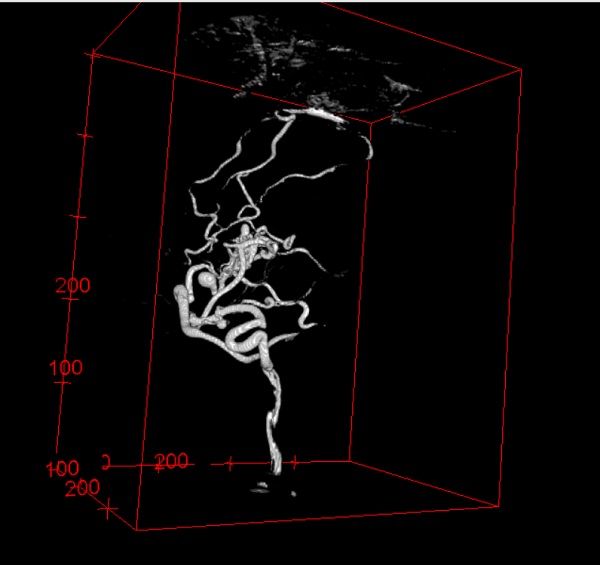
3D Dyna CT image segmentation result of AVM patient
geshi={'*.dcm','Dicom image (*.dcm)';...
'*.bmp','Bitmap image (*.bmp)';...
'*.jpg','JPEG image (*.jpg)';...
'*.*','All Files (*.*)'};
[FileName FilePath]=uigetfile(geshi,'Load MR files','*.dcm','MultiSelect','on');
if ~isequal([FileName,FilePath],[0,0]);
FileFullName=strcat(FilePath,FileName);
if ~ischar(FileFullName)
FileFullName=FileFullName([2:end 1])';
end
else
return;
end
MRo=[];
MR=[];
n=length(FileFullName);
for i=1:n
I=dicomread(FileFullName{i});
metadata=dicominfo(FileFullName{i});
serialno=metadata.InstanceNumber;
%I=rgb2gray(I);
%
MRo(:,:,serialno)=I;
MR(:,:,serialno)=im2bw(imadjust(mat2gray(I),[],[]));
end
%calculate the mask of outside bone
geshi={'*.dcm','Dicom image (*.dcm)';...
'*.bmp','Bitmap image (*.bmp)';...
'*.jpg','JPEG image (*.jpg)';...
'*.*','All Files (*.*)'};
[FileName FilePath]=uigetfile(geshi,'Load DSA files','*.dcm','MultiSelect','on');
if ~isequal([FileName,FilePath],[0,0]);
FileFullName=strcat(FilePath,FileName);
if ~ischar(FileFullName)
FileFullName=FileFullName([2:end 1])';
end
else
return;
end
DSAo=[];
DSA=[];
n=length(FileFullName);
for i=1:n
I=dicomread(FileFullName{i});
metadata=dicominfo(FileFullName{i});
serialno=metadata.InstanceNumber;
%I=rgb2gray(I);
%
DSAo(:,:,serialno)=I;
DSA(:,:,serialno)=im2bw(imadjust(mat2gray(I),[],[]));
endCode also available in Github: https://github.com/clatfd/avm/blob/remote/find_vessel.m
This is a part of my Capstone Research.