Carotid artery atherosclerotic lesion identification using a fast vessel wall imaging technique (3D-MERGE) and an artificial intelligence analysis (LATTE) technique
Background
Carotid atherosclerosis is a major cause of stroke and is associated with high mortality and morbidity globally. MRI vessel wall imaging is able to identify high-risk atherosclerotic lesions and may improve diagnosis sensitivity and accuracy. Traditionally, 2D high-resolution multi-contrast MRI (2D-MC MRI) is the most reliable and reproducible noninvasive carotid vessel wall imaging technique. And manual review on MRI by experoienced radiologists is the usual practice. However, the long scan time required for 2D-MC MRI and the lack of trained radiologists with extensive experience in the evaluation of carotid plaque morphology and composition restrict the practical clinical application of MR vessel wall imaging.
For the imaging side, our group developed a fast 3D carotid black blood MRI sequence (3D Motion Sensitized Driven Equilibrium prepared Rapid Gradient Echo, 3D-MERGE) [Balu N, MRM 2011], which allows large coverage of carotid arteries with submillimeter isotropic resolution in coronal acquisition, and is able to depict atherosclerotic lesion burden, severity, and luminal stenosis. This rapid sequence can complete a carotid scan in 2 min.
However, for the image analysis side, the image review for 3D-MERGE is even more complex due to its single sequence available and the much larger coverage (number of slices) required for reading. Artificial intelligence (AI) techniques might be the solution to the image analysis challenge. However, for this lesion classification task, we might likely face the problem of domain shift, meaning the trained model might not work well on new unseen test dataset during deployment, even the testing dataset is only slightly different in different coils, imaging parameters, scanner hardware and software releases.
Aims
1. Feasibility
To use 3D-MERGE as a alterative for traditional 2D-MC MRI, we need to prove that 3D-MERGE alone can serve as a screening tool to identify advanced carotid lesions.
2. Training a lesion classification model is easy, but we need to consider three important factors so that the model can be clinically usable.
2.1 Scan quality check
MRI was easily affected by patient's motion. When the image quality is low, the whole 3D scan is not diagnosable. It can save a lot of trouble if we can inform the MR operator to rescan the patient before the patient leaves the scanner.
2.2 Automation
Only a fully automated solution has the potential to be a screening tool for batch process on large number of cases. Thus we need to solve the problem of artery localization (where are the arteries in 3D space) and identification (which is the target artery of interest)
2.3 Domain adaptive
Only a domain adaptive solution can be deployed in hospitals with different scaners. Ideally the adaptation is unsupervised, meaning no additional annotations needed to the new sites.
Solutions
Feasibility
In the [Stroke paper], we used 2D-MC MRI to identify advanced lesions (plaques of AHA type IV and above) from 449 arteries/227 participants as the reference. We compared the diagnosis ability to identify advanced lesions from 3D-MERGE alone. Results show the sensitivity, specificity, and accuracy for identification of advanced lesions on 3D-MERGE were 95.0% (95% CI: 91.8 - 97.2), 86.9% (95% CI: 81.4 – 92.0), 93.8%(95% CI: 91.1 -95.8) respectively, a good agreement with 2D-MC, but 3D-MERGE saves much time in scanning and covers more area (more likely to find more lesions otherwise ignored).
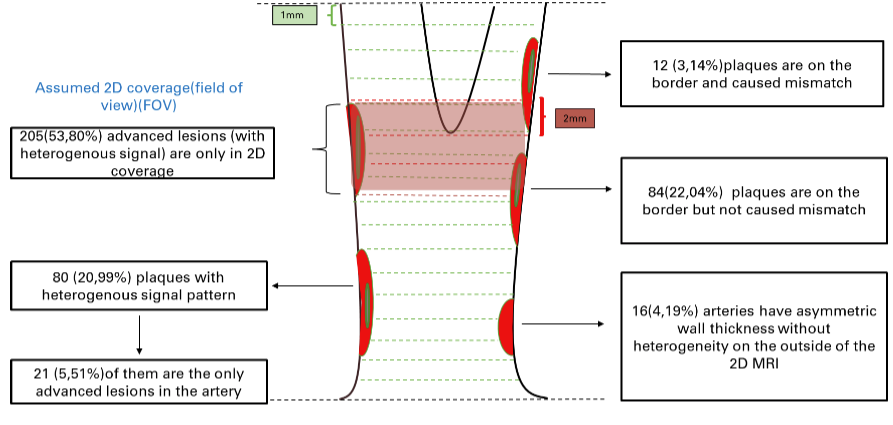
Illustration of plaque distribution on 3D-MERGE
Image quality assessment
In the [SPIE paper], we developed an image quality assessment tool, which can predict the image quality for all axial slices. The solution is a target-oriented patch based deep learning algorithm for medical image quality assessment. We first used an artery detection neural network (built on YOLOv2) to identify bounding boxes of artery locations, then crop and split each axial slice to grids of patches around the artery region. Each patch predicted an image quality score and each patch was set with weights according to the distance to the detected artery location. Finally, the image score is determined by a weighted average of image quality scores from each patch.
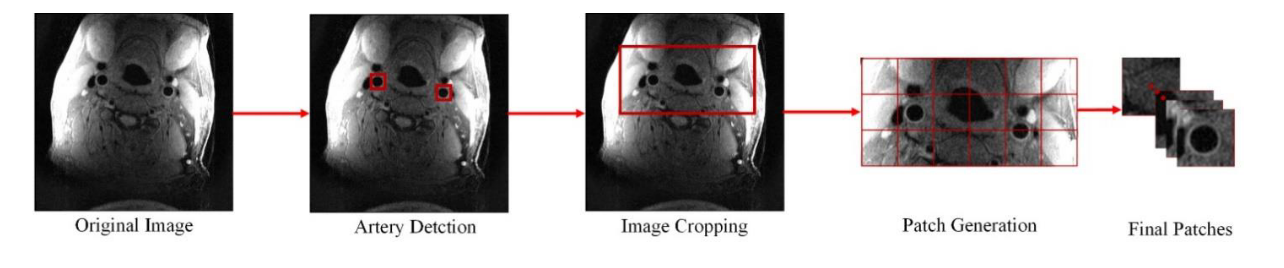
Pipeline for generating patches from the original image
With all the slices predicted with a quality score, we can decide whether the whole scan is with poor quality. A rescan notice can be given at this time. Otherwise, we can further select the smaller range of axial slices for further analysis, as the begining and ending axial slices for a carotid scan are likely to contain little information (coil receives little signal).
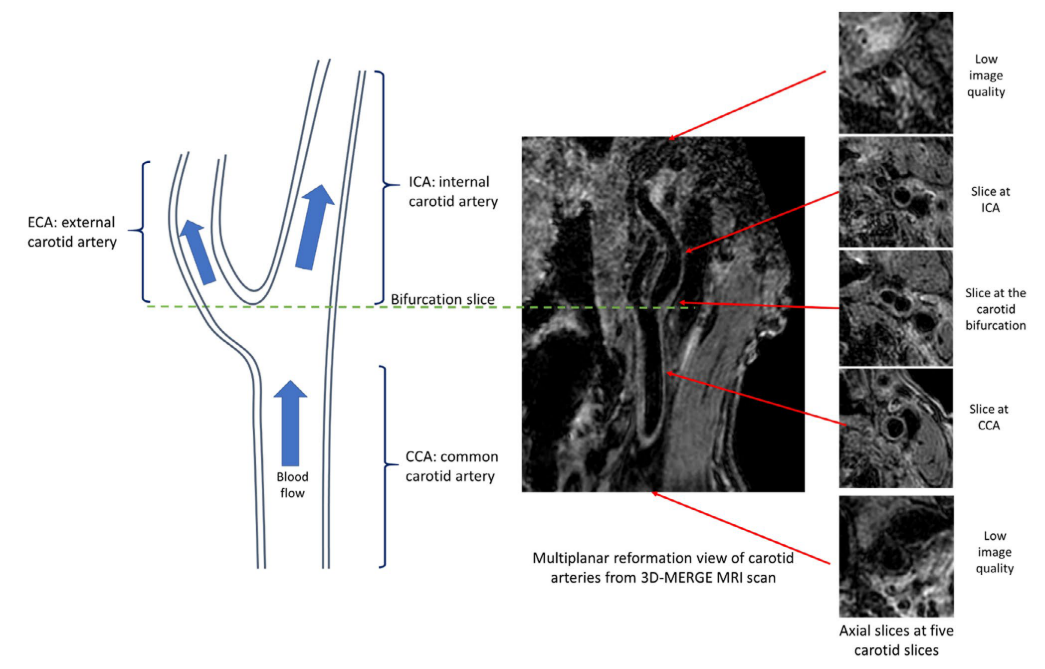
Illustration of carotid anatomical regions with example 3D-MERGE images of carotid arteries. Slices at both ends have low image qualities; thus, the image quality assessment module is used to predict the qualified slices for further analysis
Automation
The task is very similar to FRAPPE, where the artery is realtively straight and the spacing between axial slices are reasonable. Like FRAPPE, we continue to use the tracklet refinement appoach. First aggregate closely overlapping artery detections in each slice (same Yolov2 results in image quality assessment) to tracklets, then merge tracklets into centerlines according to 3D neighboring information. Finally we select up to 4 main arteries centerlines (2 on each side of neck) and label them with external carotid artery (ECA), internal carotid artery (ICA) or common carotid artery (CCA) according to their relative locations.
Patches along the centerlines were extracted, then vessel wall region were segmented (to highlight analysis region) for lesion classification.
Domain adaptation
Detailed in the [MRM paper], we proposed an unsupervised domain adaptive solution for lesion classification. Here we classify patches (vessel wall status at certain location along the centerline) into three categories for labeling: -1 (normal artery), 0 (early lesion), and 1 (advanced lesion)
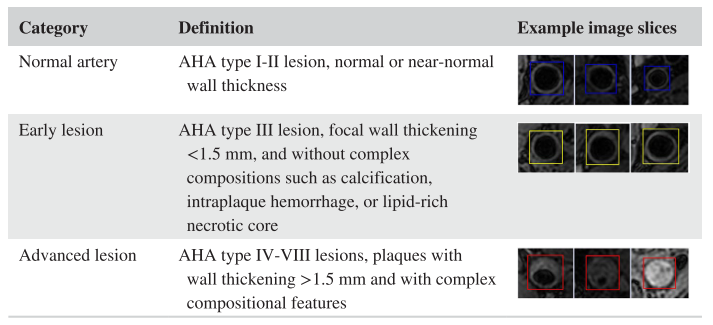
Carotid vessel wall categories with definition and example image slices
Our domain adaptive solution is able to transfer the lesion classifier trained from the source domain (with manual annotations classifying lesion types) to new target domains without additional manual annotations.
In addition to the source classifier branch S(P), where the artery at the center slice was classified into three categories y, a domain classifier branch D(P) was added after the last convolutional layer to discriminate the domain of the inputs (source or target).
An alternate procedure was used in training the network.
In step 1, weights of the domain classifier branch were fixed for training the rest of the network using both patches from the source Ps,i and target domains Pt,i. The loss of step 1 (l1) was the sum of the source classifier loss and the domain classifier loss. The source classifier loss is the mean absolute error loss from the source classifier branch where the ground truth category of the center patch of the input ys,i was compared with the predicted category S(Ps,i). The domain classifier loss is the binary cross entropy loss from the domain classifier branch where the ground truth domain ds/t,i was compared with the predicted domain D(Ps/t,i). It should be noted that the source classifier did not require labels from patches of the target domain (yt,i set to 0) and the loss calculation of the source classifier did not include target domain patches due to the unsupervised DA design. i indicates each patch in the source or target domain.
In step 2, the weights other than the domain classifier were fixed, and the same input was used to train the domain classifier branch only. Importantly, an adversarial approach was used here to invert the domain labels on purpose to train the model, so that if the source classifier was robust enough, it could not tell whether the features extracted from the con- volutional layers were originated from the source or target domain. In other words, the convolutional layers were trained to extract only features that were shared across different domains to make a correct classification in the source classifier. The loss in step 2 (l2) was only the binary cross entropy loss from the domain classifier branch.
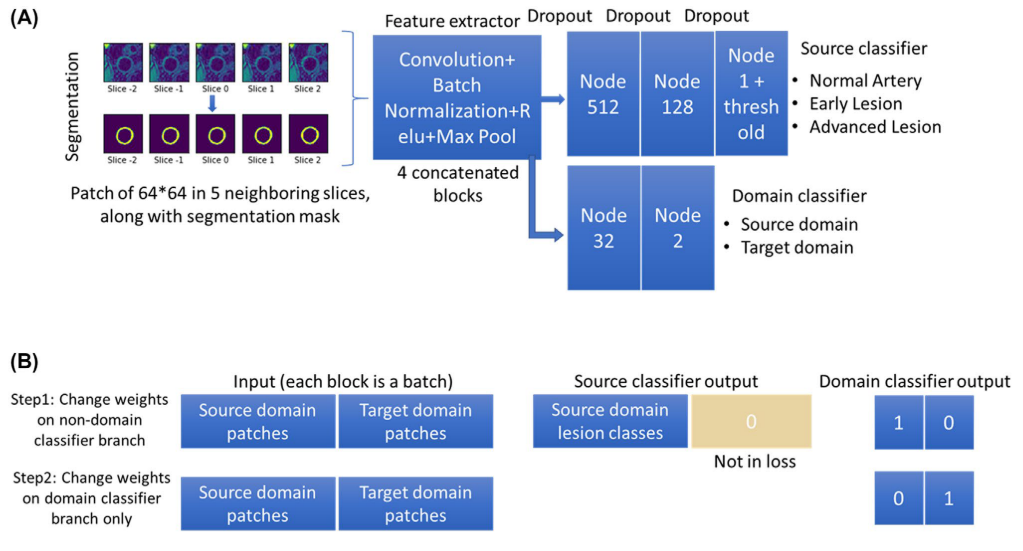
Network structure and training stretegy for the domain adaptive lesion classifier
LATTE
Now we have the global picture of LATTE [Lesion Assessment Through Tracklet Evaluation], a solution for carotid artery atherosclerotic lesion imaging and analysis.
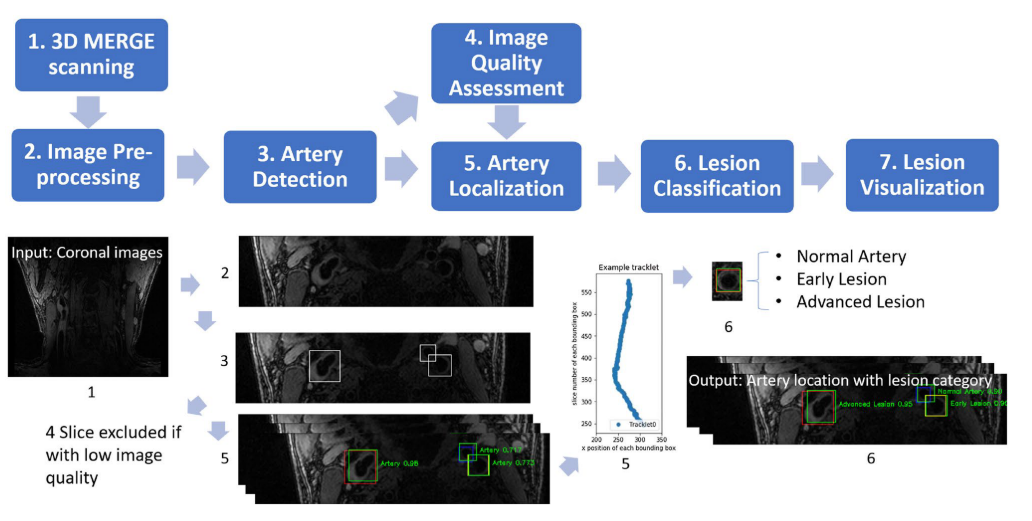
Workflow for LATTE
Some of the results
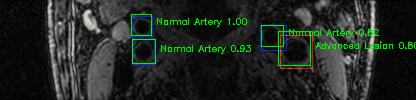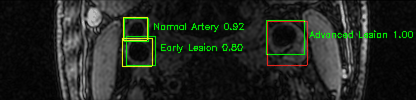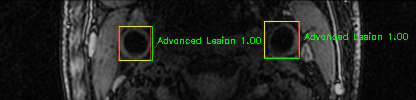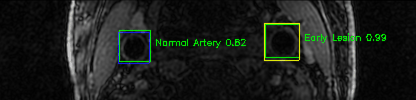
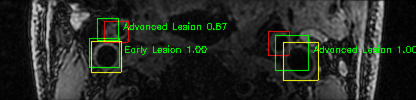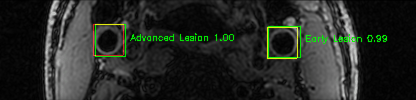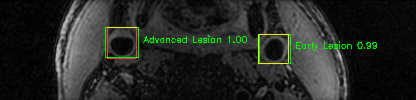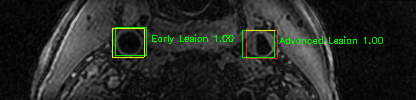
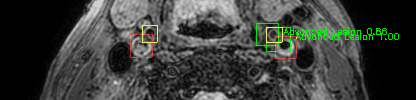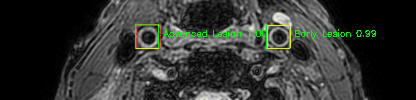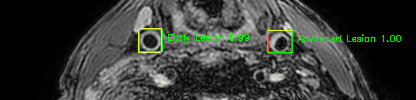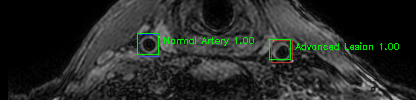
Green bounding boxes: predictions with lesion type and confidence
Blue/yellow/red bounding boxes: ground truth labels for normal/early lesions/advanced lesions
Relation with iCafe family
Artery identification part is using the similar tracklet refinement algorithm with FRAPPE.
Results can be exported as CASCADE format, so that vessel wall segmentation results can be manually modifed then vessel wall features can be quantified/analyzed.
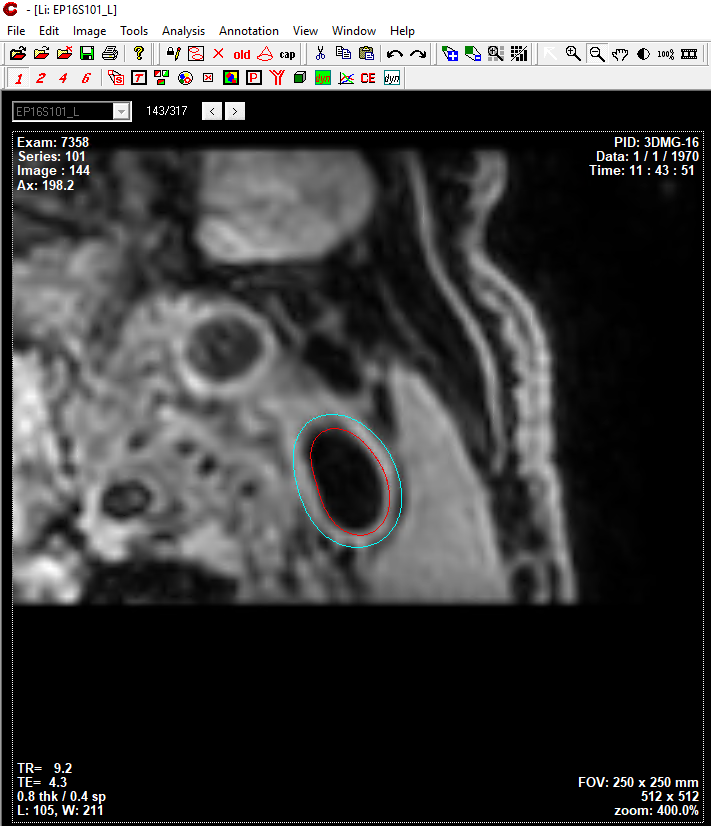
Segmentation results shown in CASCADE
Results can be exported as the iCafe project format, so that results can be viewed in iCafe.
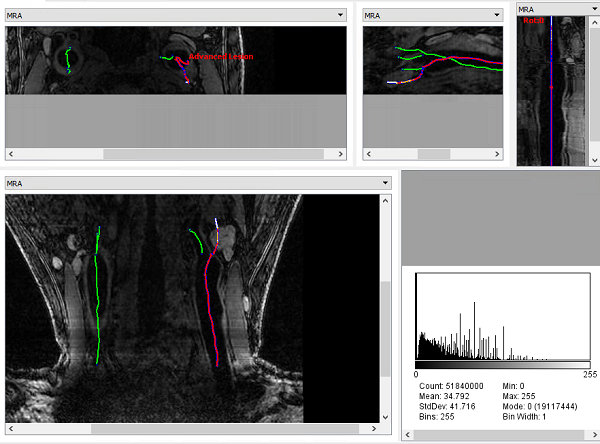
iCafe view (axia, sagittal, multiplanar reformation, coronal view, histogram) of LATTE results
Related publications
Duygu Baylam Geleri, Hiroko Watase, Baocheng Chu, Li Chen, Huilin Zhao, Xihai Zhao, Thomas Hatsukami, and Chun Yuan. Detection of Advanced Lesions on Carotid Arteries using 3D-MERGE Magnetic Resonance Imaging as a Screening Tool. Stroke (IF:7.2), 2021, Accepted.
Li Chen, Huilin Zhao, Hongjian Jiang, Niranjan Balu, Hiroko Watase, Duygu Baylam Geleri, Xihai Zhao, Rui Li, Jianrong Xu, Thomas S. Hatsukami, Dongxiang Xu, Jenq-Neng Hwang, Chun Yuan. Domain Adaptive and Fully Automated Carotid Artery Atherosclerotic Lesion Detection using an Artificial Intelligence Approach (LATTE) on 3D MRI. Magnetic resonance in medicine (IF:3.6), 2021, Accepted. DOI: 10.1002/MRM.28794
Hongjian Jiang, Li Chen, Dongxiang Xu, Huilin Zhao, Hiroko Watase, Xihai Zhao, Rui Li, Chun Yuan. A Target-Oriented and Multi-Patch Based Framework for Image Quality Assessment on Carotid Artery MRI. Medical Imaging 2020: Image Processing. SPIE, 2020, Marriott Marquis Houston, Houston, Texas, United States (February 15 - 20). DOI: 10.1117/12.2549473
Li Chen, Huilin Zhao, Niranjan Balu, Xihai Zhao, Rui Li, Jianrong Xu, Thomas S Hatsukami, Jenq-Neng Hwang, Chun Yuan. Carotid Artery Localization and Lesion Classification on 3D-MERGE MRI using Neural Network and Object Tracking methods. 2019 Annual Meeting of International Society for Magnetic Resonance in Medicine (ISMRM), 2019, Palais des congrès de Montréal, Montréal, QC, Canada (May 11-16). (pdf) (ppt)
Li Chen, Huilin Zhao, Niranjan Balu, Xihai Zhao, Rui Li, Jenq-Neng Hwang, Chun Yuan. Carotid Artery Localization and Lesion Detection on 3D-MERGE MRI through Online Learning.. Society for Magnetic Resonance Angiography 30th Annual International Conference, 2018, University of Glasgow, Glasgow, Scotland (August 29-31).