Fully automated and Robust Analysis Technique for Popliteal Artery Vessel Wall Evaluation (FRAPPE)
A 7-minute presentation by Li Chen (during Imaging Elevated symposium 2020): https://youtu.be/2U5saNmYlzo
10-slide PPT http://clatfd.cn/data/FRAPPE10.pdf
Based on our observations [paper], vessel wall structures in routine MR Knee scans are visible and analyzable. Quantitative analysis of popliteal vessel wall can provide additional risk scores for assessing cardiovascular risks, as atherosclerosis is known to be a systemic disease. The OAI dataset with more than 3.5 million knee images is a valuable dataset for knee scans, but sadly, beyond knee and osteoarthritis analyis, nobody has paid attention to the popliteal vessel wall.
So our aim is to develop a tool which can accurately locate popliteal artery from knee images and quantify vessel wall features in a fully automated way.
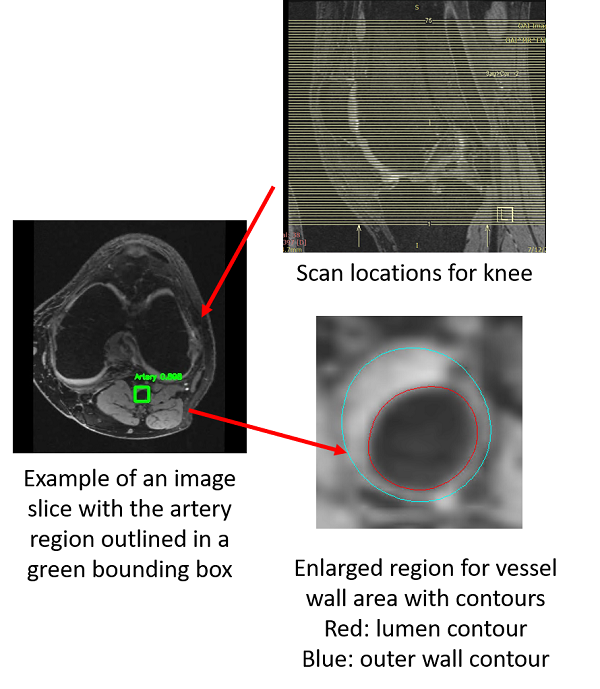
However, there are many challenges ahead.
1. Find artery locations from 3D MRI images is not easy. Pathological changes affecting detection, neighboring information need to be used, etc.
2. Segment vessel wall accurately is challenging. Noise, artifacts, coil problems, etc. Deep learning might be useful.
3. Train a deep learning model with few annotations. Manual labeling vessel wall in each slice of 3D knee scans takes more than 3 hours per scan.
So we developed the FRAPPE (Fully automated and Robust Analysis Technique for Popliteal Artery Vessel Wall Evaluation) workflow shown in the figure below. More details are available in our MRM paper.
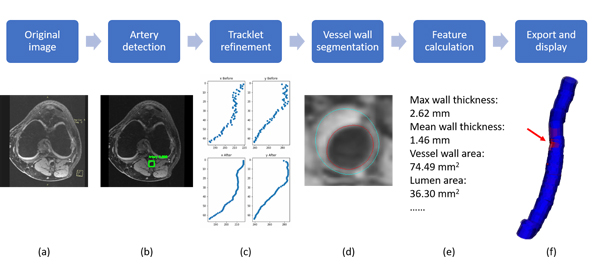
FRAPPE workflow
Technical details
Five steps in FRAPPE
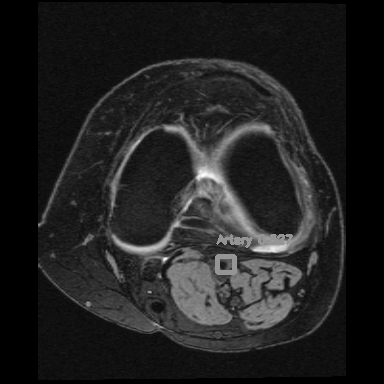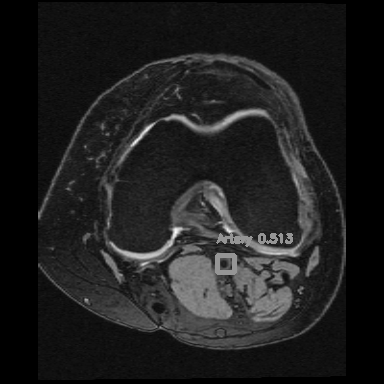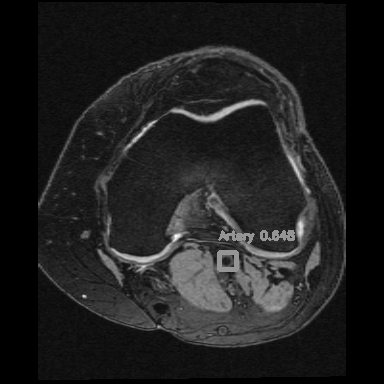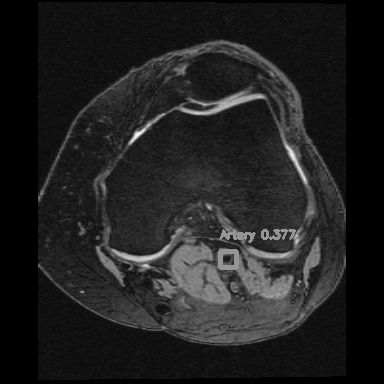
Popliteal artery locations were detected from each MR slice by a deep neural network model (YoloV2).
The detection might not be perfect (with additional detections and missing detections), so we propose to connected detections in each slice into a 3D artery centerline by tracklet refinement (arXiv).
Vessel wall regions around the centerline were then segmented using another neural network model for segmentation in polar coordinate system.
Contours from vessel wall segmentations were used for vascular feature calculation, such as mean wall thickness and wall area.
Finally, results can be exported in iCafe for color coded 3D visualization.
If you consider the time axis as the depth axis in 3D images, detecting arteries is equivalent to detecting objects in a video.
 <=>
<=>
Left: Car detections from Z. Tang, CVPR, 2018
Right: Artery detection in knee scan
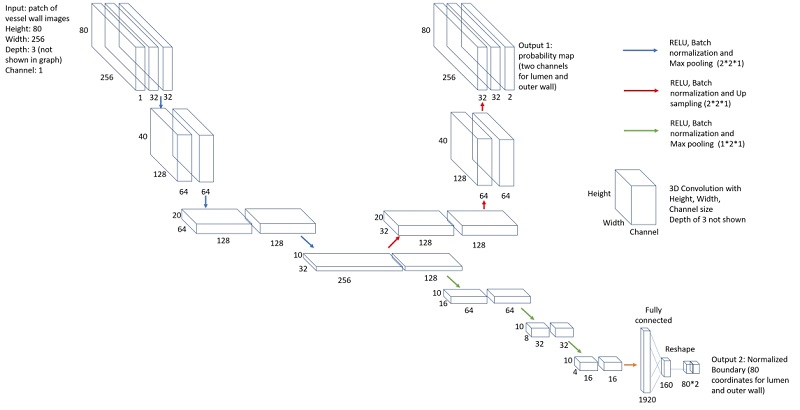
The network structure for vessel wall segmentation
We used transfer learning approach to train the vessel wall segmentation from the model on carotid arteries, as we have more abundant carotid vessel wall contours. This can reduce the number of cases required for training.
In addition, to cope with challenging slices, for example, many popliteal vessel wall slices have calcified plaques, which are not so common in carotid vessel wall, we used the active learning idea. We selected the cases with poor performance and asked human reviewers to correct the mistakes, then the new contours are combined with the previous training set to further train the model.
The below examples show the improvements on the testing set after active learning approaches.
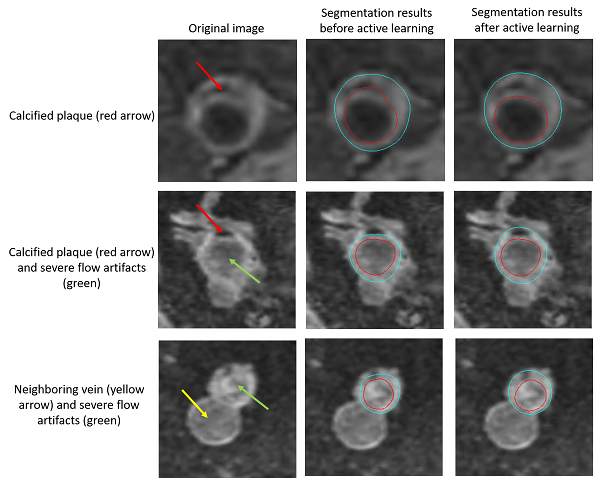
Improvements on vessel wall segmentation after active learning approaches
This new popliteal artery analysis technique (FRAPPE) was validated against manual segmentation qualitatively and quantitatively in a series of 225 cases from the Osteoarthritis Initiative dataset. FRAPPE demonstrated high accuracy and robustness in locating popliteal arteries, segmenting artery walls and quantifying arterial features. Qualitative evaluations showed 1.2% of slices had noticeable major errors, including segmenting wrong target and irregular vessel wall contours. The mean Dice similarity coefficient with manual segmentation was 0.79, which is comparable to inter-rater variations. Repeatability evaluations show most of the vascular features have good to excellent repeatability from repeated scans of same subjects, with intraclass coefficient ranging from 0.80 to 0.98.
The below examples show the challenging slices with bifurcation, low contrast, large plaque or vein co-existing.
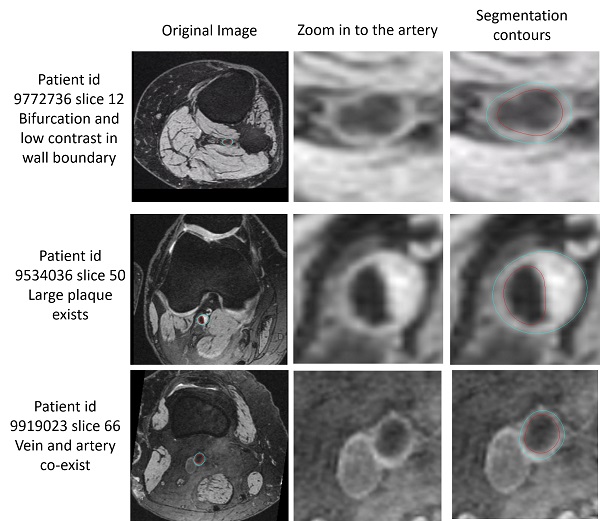
Challenging slices while FRAPPE still acheives good results
Based on our workstation with Intel Xeon CPU E5-1650 v4 @3.6GHz 6 cores, 64 GB memory, NVIDIA GeForce GTX TITAN V on Windows 10, the processing time for Testing Set 2 for artery localization, vessel wall segmentation and feature calculation is 65.2±19.8 seconds, 279.1±48.4 seconds, and 114.3±14.7 seconds, respectively. The whole process took less than 8 minutes per artery on average. We have processed all the cases in two months using 7 GPUs working in parallel.
This project won the first place in the American Heart Association/Amazon Web Services Prize Competition. (News)
The contours for popliteal vessel wall are shared in https://github.com/clatfd/OAI-Polar
Technical paper published in Magnetic Resonance in Medicine
Media reports:
https://newsroom.heart.org/news/seattle-scientist-wins-competition-for-artificial-intelligence-research
https://newsroom.uw.edu/postscript/knee-mris-can-depict-cardiovascular-risk-ai-affirms-it
https://www.ece.uw.edu/spotlight/chen-knee-mri/
Editorial from Dr. David A. Bluemke on JAHA
Manual labels made public
The manual contours used for development of FRAPPE is made public at https://github.com/clatfd/OAI-Polar
Follow up research
During the technical development, the vessel wall features are showing predictive values for subjects with high cardiovascular risks. So we believe there is valuable information if we can quantify vessel wall and use that as biomarkers.
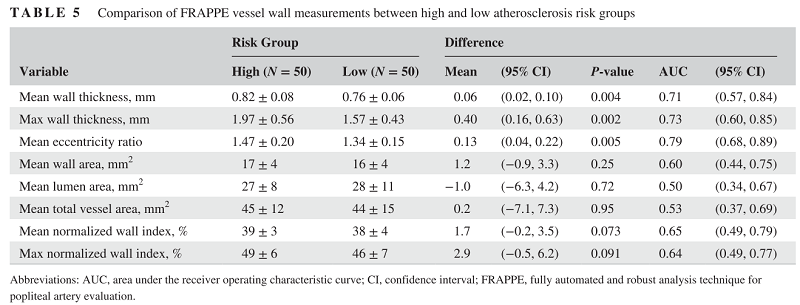
With all the contours segmented, and clinical information in the OAI dataset, we can do a lot of follow up medical research.
Some on-going projects include:
Plaque identification through feature map analysis ![]() ISMRM2020_plaque.pdf
ISMRM2020_plaque.pdf
Lumen and vessel wall remodeling pattern (JAHA ![]() JAHA.120.018408.pdf)
JAHA.120.018408.pdf)
Relation between vessel wall and physical exercise (under review)
Relation of vessel wall and diseases (diabetes, heart attack)