This is a part of my Capstone Research.
The benefit of using graph network to describe brain vessel is its plasticity. The vessel shape may slightly change in each imaging modality and its orientation or location may be different. Using network to describe the image means using nodes and links instead of voxel to determine the location and connectivity of vessels, avoiding many problems in different modalities.
Main procedure:
1. Slicing in Z index
Select vessel regions in each slice of picture. Starting from the top slice with Z=0. Calculate area of each vessel area, record the location and area(as node weight).

four vessels in this slice
2. Connectivity measure
If there are also vessel(s) in the area of vessels from last slice, assume these two nodes are connected. Record the connections.
3. Trim connection
After these two steps, there are more than 3000 nodes and 2000 connections. That is a large number for further calculation. So the trimming is required.
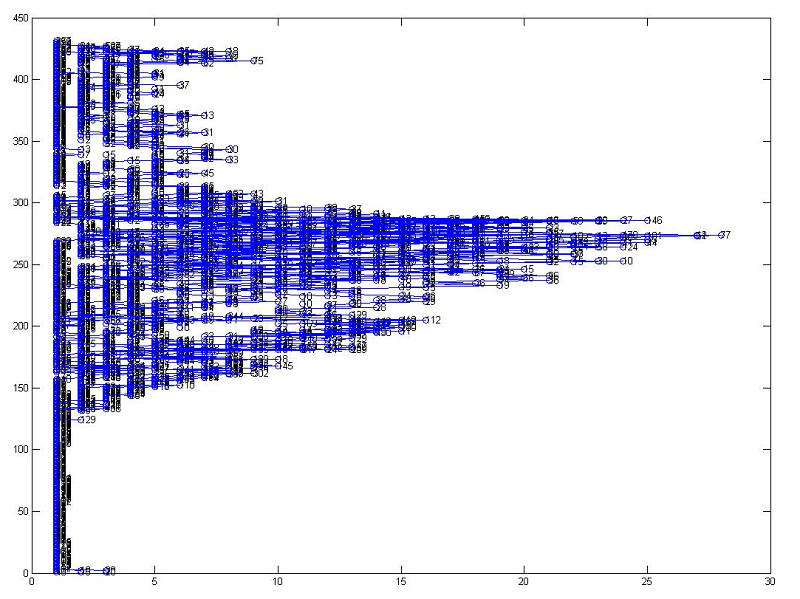
Too many nodes and connections for calculation(numbers in circles are weight of nodes)
The basic idea of trimming is to delete unnecessary nodes and connections. First, if node area is too small(less than 10 pixels), these are tiny vessels that can be ignored. Second, if the node is following a same direction without divergence, these nodes can be combined in a heavy-weight node representing a large vessel.
After trimming, the graph is much simpler.
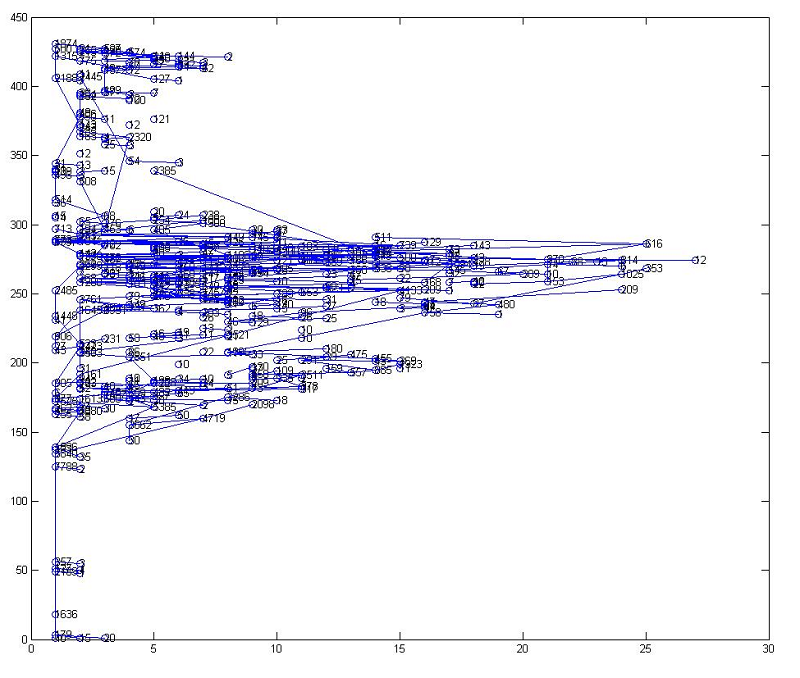
After trimming
Matlab code available in github: https://github.com/clatfd/avm
net_raw.m
net_trim.m
.