The data is 3D TOF MRA image for carotid vessel. In TOF method, vessels are much brighter than other tissue, so using a thresholding method is enough to perform segmentation.
For a more convenient way to choose an appropriate threshold value and cropping region, a Matlab GUI is designed to select value with slider and on-picture clicking. Move the slider on the left, then thresholding result is displayed on the right. Click on the right picture twice to determine the left-top and right-bottom value for cropping.
To remove other small obstacles in the 3D picture, connectivity of 3D region is used to label parts in the image. If the part is below a certain minimum voxel, it is removed.
Output image stack and view picture in ImageJ.
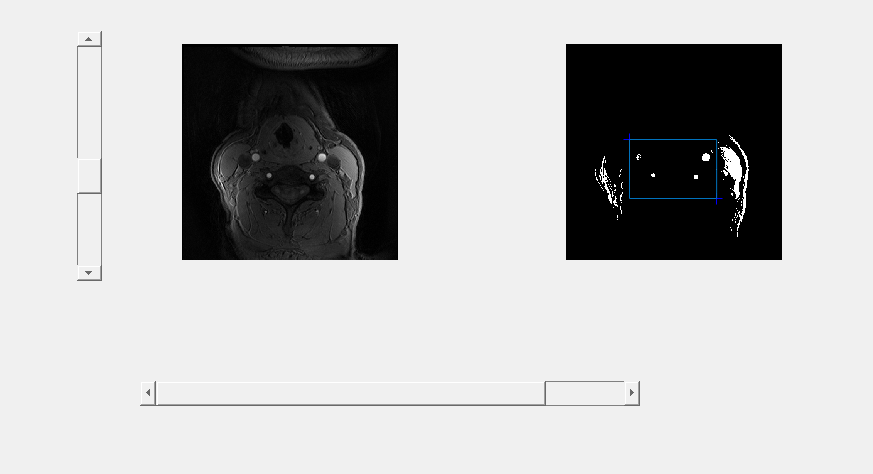
GUI for value selection
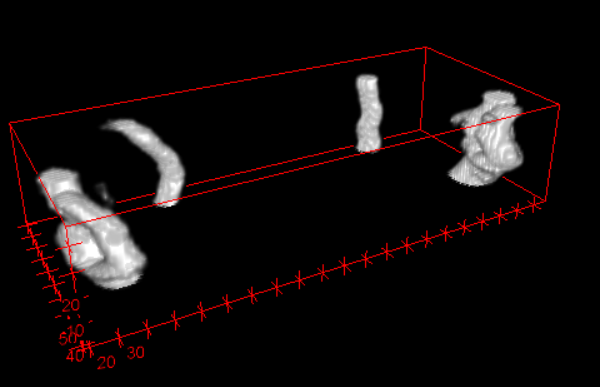
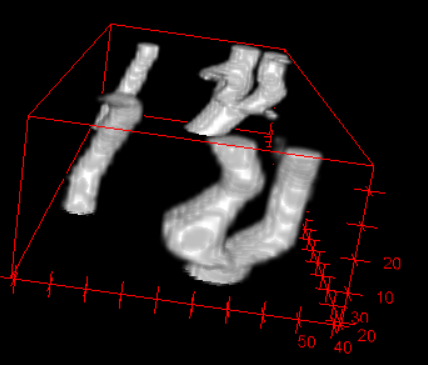
3D view for segmentation result
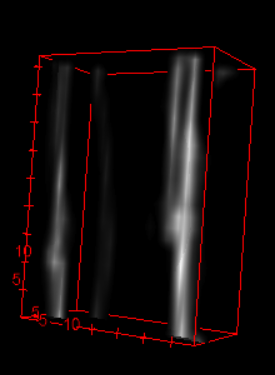
Resize to fit thickness
Main.m
%Matlab code for segmentation of 3D carotid vessel by thresholding to a 3D %TOF image %Author: Li Chen (http://clatfd.cn) D=load3dtof(); %Or >> load data_ori.mat %GUI for adjusting threshold in im2bw, %and selecting two points(first is left-top corner, second is right-bottom %corner) to crop, then close windows(variable stored automatically) sliderimg(D); Da=D(crop(2):crop(4),crop(1):crop(3),:); %crop Db=im2bw3D(Da,thres); outtotif(Db,'output_tif'); %output to tif files in folder %save Db 'output_crop.mat'; Dc=rmsmallobj(Db,500); %remove small(voxel < 500) and seperated part outtotif(Dc,'output_tif_r'); %save Dc 'output_cr.mat';
Code:
Github:
https://github.com/clatfd/ThresSeg3d